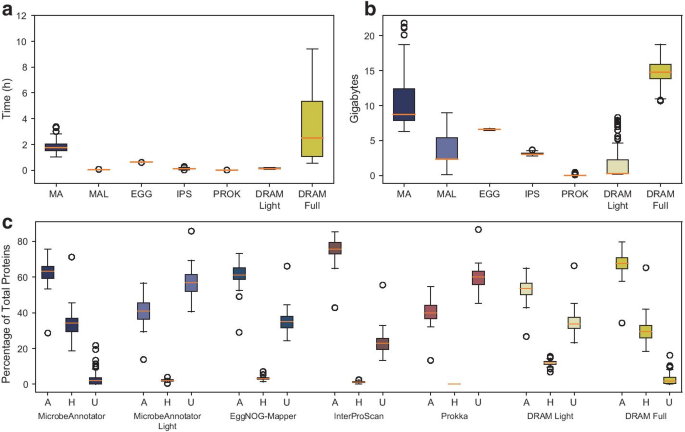
MicrobeAnnotator: a user-friendly, comprehensive functional annotation pipeline for microbial genomes | BMC Bioinformatics | Full Text
Prokka: NCBI prokaryotic genome submission, mobile element re-annotation & tbl2asn · Issue #272 · tseemann/prokka · GitHub

Genomic Analysis of the Evolution and Global Spread of Hyper-invasive Meningococcal Lineage 5 - ScienceDirect

Prokka genome annotation coverage of Firmicutes (GTDB taxonomy) against... | Download Scientific Diagram

INfrastructure for a PHAge REference Database: Identification of large-scale biases in the current collection of phage genomes | bioRxiv
![Whole genome sequencing of a novel, dichloromethane-fermenting Peptococcaceae from an enrichment culture [PeerJ] Whole genome sequencing of a novel, dichloromethane-fermenting Peptococcaceae from an enrichment culture [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2019/7775/1/fig-1-full.png)
Whole genome sequencing of a novel, dichloromethane-fermenting Peptococcaceae from an enrichment culture [PeerJ]
Overview of the MagCluster workflow. (a) Genomes are annotated using... | Download Scientific Diagram
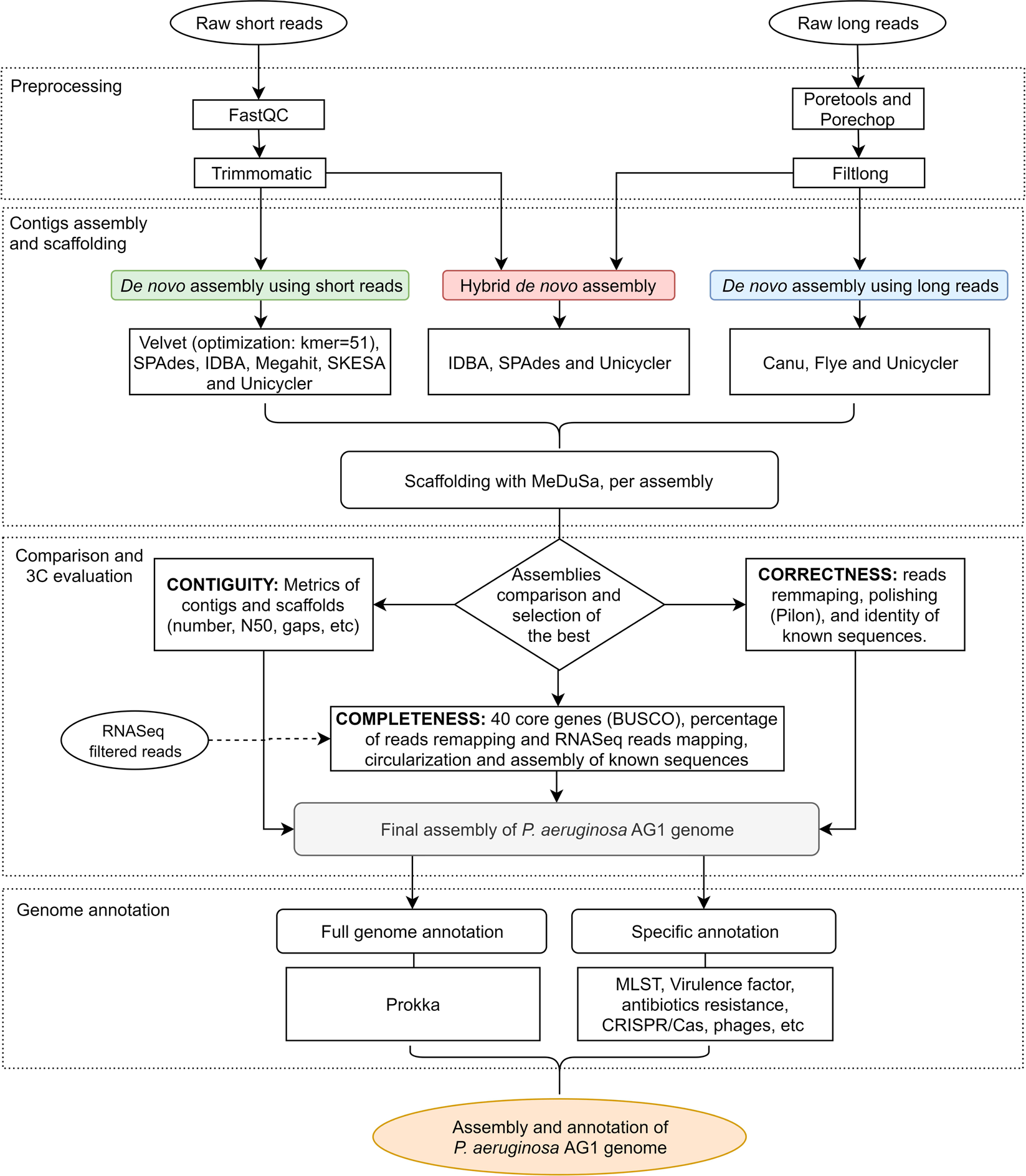
High quality 3C de novo assembly and annotation of a multidrug resistant ST-111 Pseudomonas aeruginosa genome: Benchmark of hybrid and non-hybrid assemblers | Scientific Reports

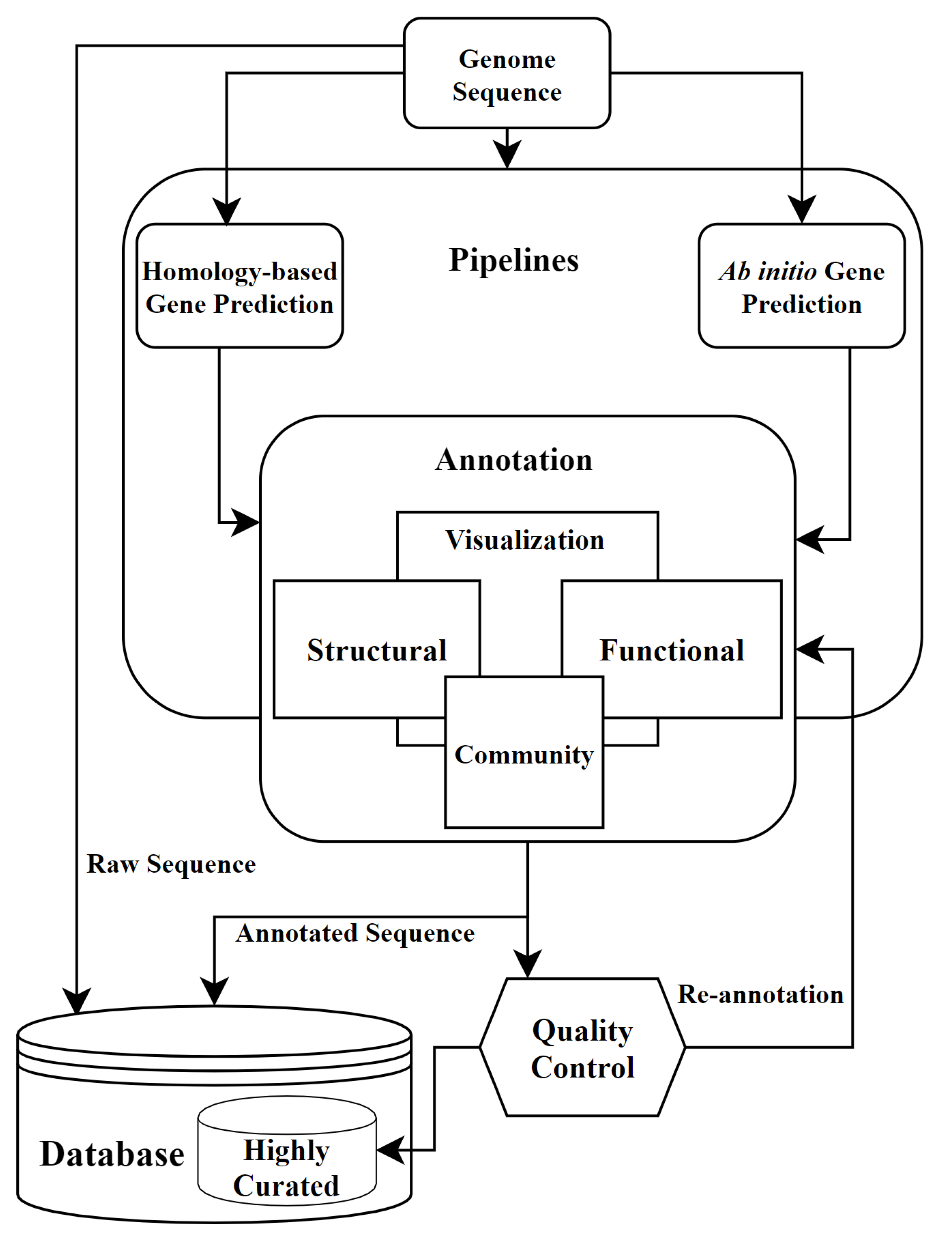
Automated analysis of genomic sequences facilitates high-throughput and comprehensive description of bacteria | ISME Communications
![PDF] Prokka: rapid prokaryotic genome annotation | Semantic Scholar PDF] Prokka: rapid prokaryotic genome annotation | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/4b57f72ae7d091863dc87b852cd01823e7644223/2-Table3-1.png)